Tutorial¶
To further demonstrate the functionality of Parsnp we have prepared two small tutorial datasets. The first dataset is a MERS coronavirus outbreak dataset involving 42 isolates. The second dataset is a selected set of 31 Streptococcus pneumoniae genomes. Both of these datasets should run on modestly equipped laptops in a few minutes.
42 MERS Coronavirus genomes
Download genomes:
Run parsnp
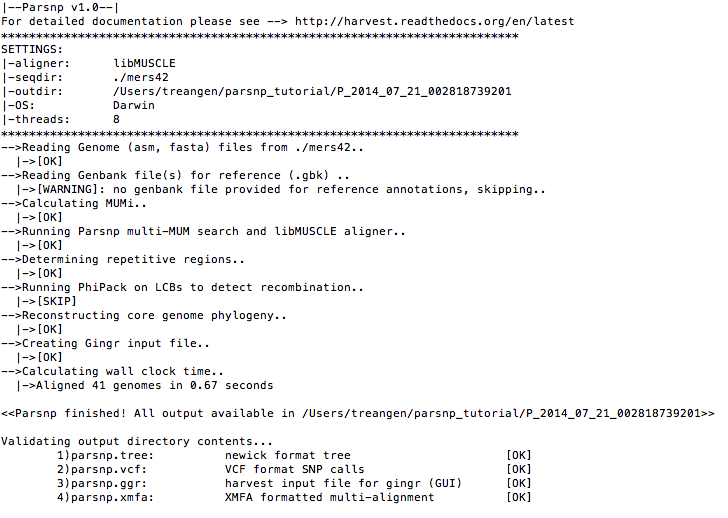
./parsnp -r ./mers42/England-Qatar_2012.fna -d ./mers42Command-line output
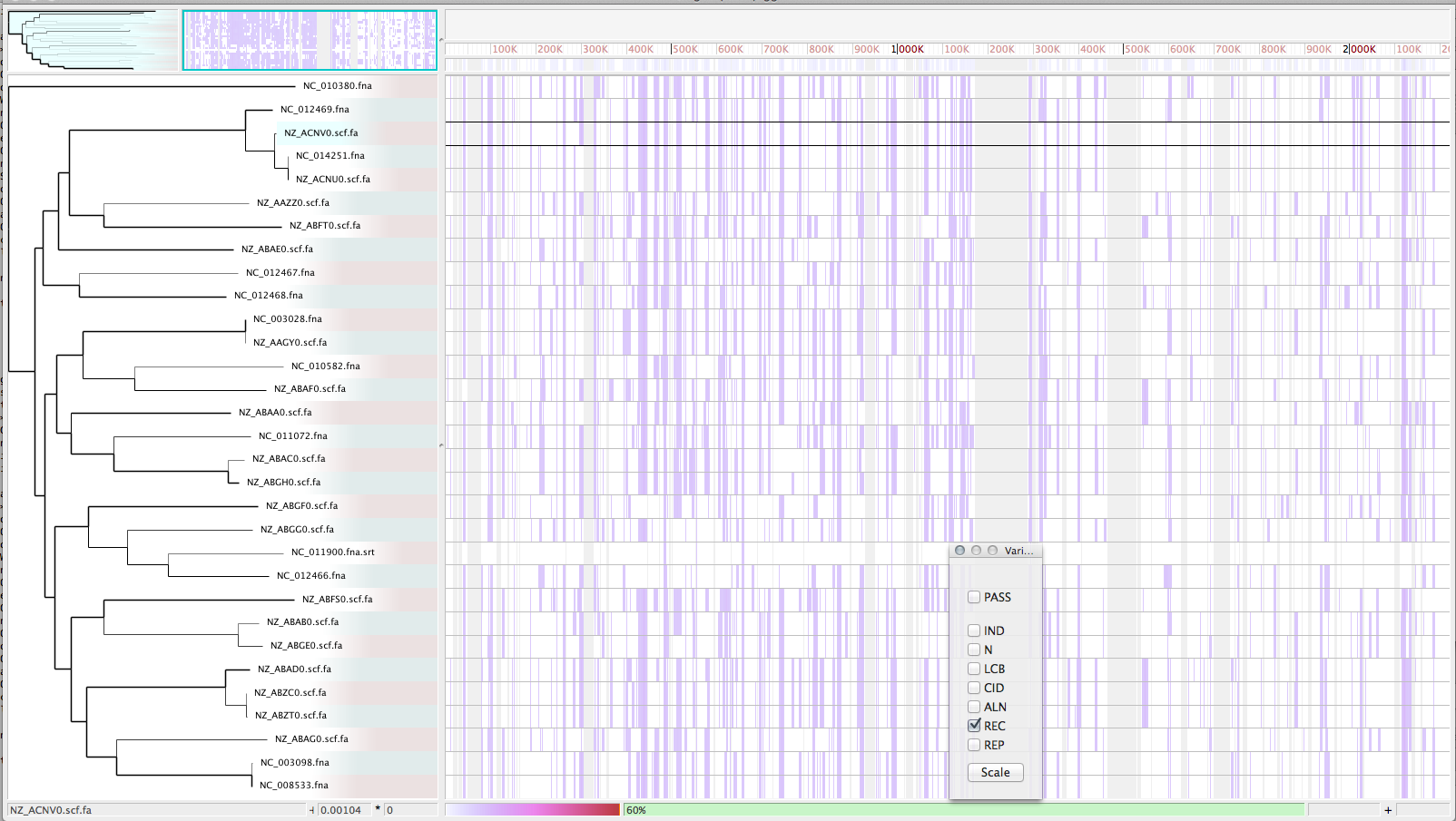
- Visualize with Gingr:
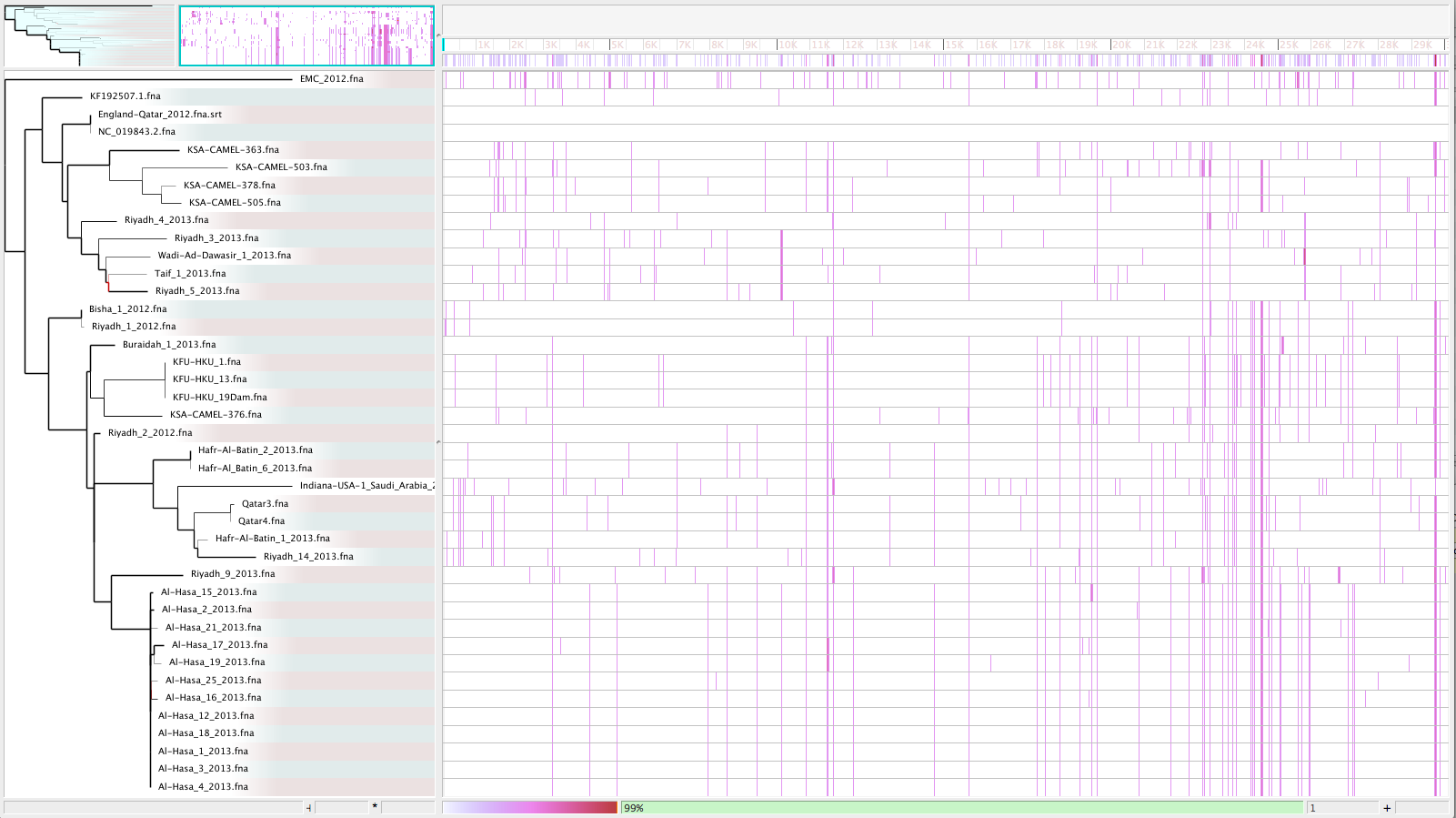
Inspect Output:
31 Streptococcus pneumoniae genomes
Download genomes:
Run parsnp
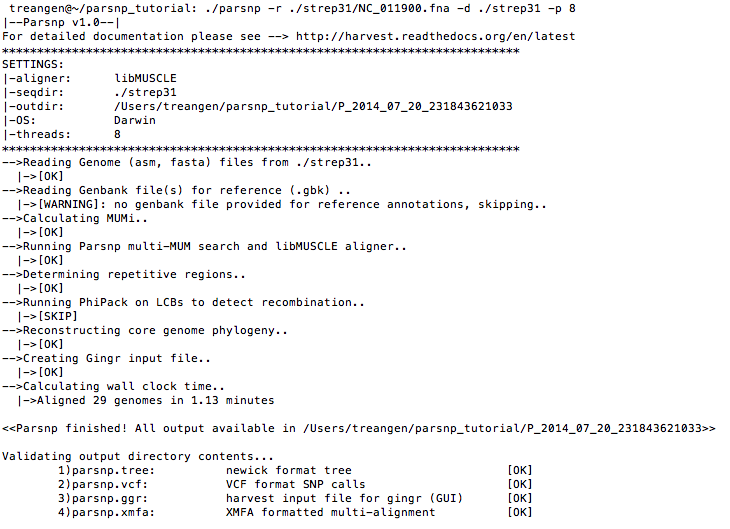
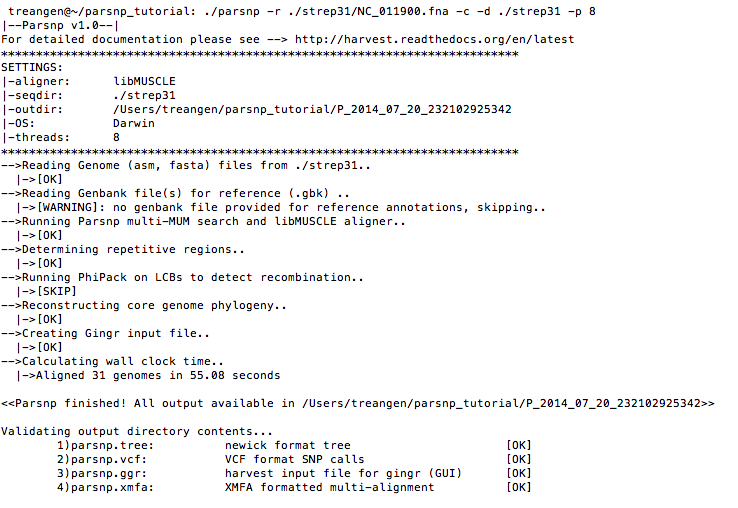
./parsnp -r ./strep31/NC_011900.fna -d ./strep31 -p <num threads>Example output:
Force inclusion of all genomes (-c)
./parsnp -r ./strep31/NC_011900.fna -d ./strep31 -p <num threads> -c
Command-line output:
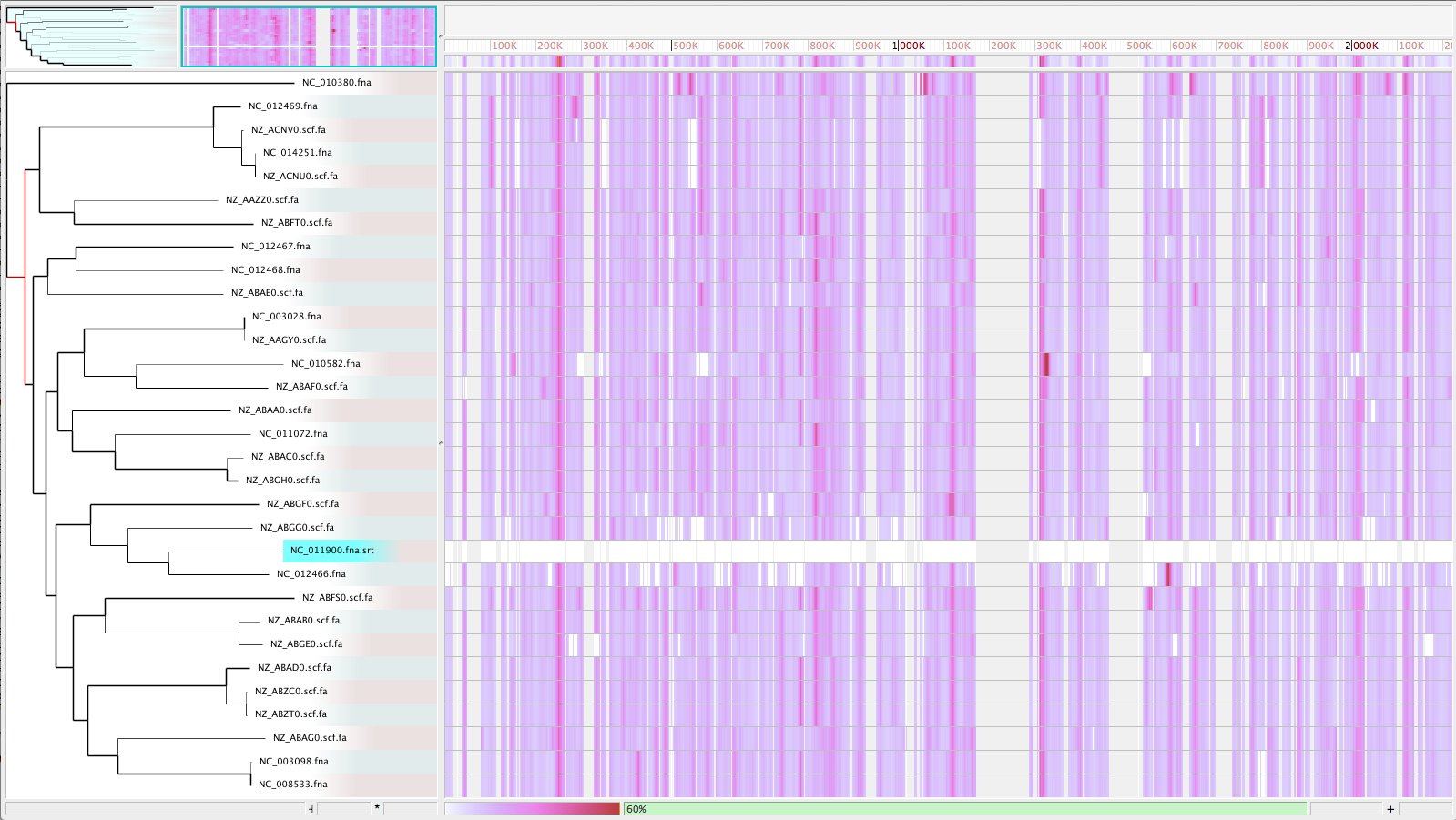
Visualize with Gingr
Enable recombination detection/filter (-x)
./parsnp -r ./strep31/NC_011900.fna -d ./strep31 -p <num threads> -c -xRe-visualize with Gingr
- Bootstrap values have improved after running recombination filter; columns with filtered SNPs are displayed in image: